hmmIBDr
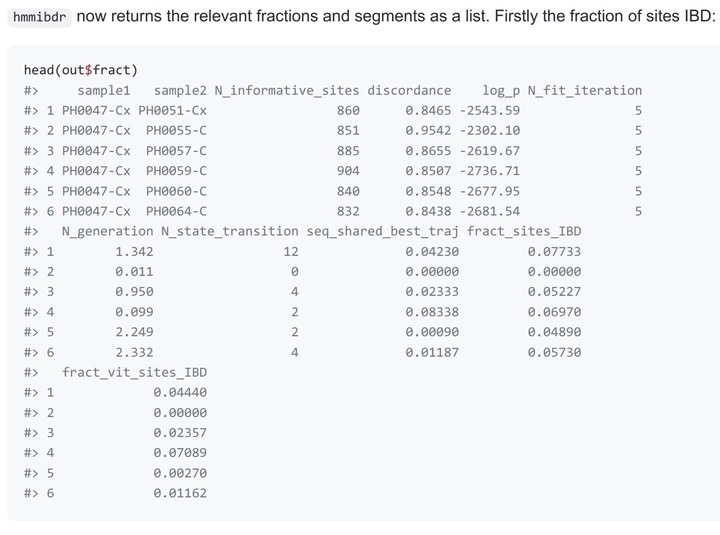
hmmibdr is an Rcpp wrapper for hmmIBD in Rcpp. There has been a recent proliferation in the number of malaria studies using IBD. As interest in IBD grows, the need to provide comprehensive details of software used to infer IBD increases. hmmIBD is the only program known to the authors that is designed specifically for haploid malaria genomes and is capable of comparing samples across populations with different allele frequencies. This will likely be a useful feature for malaria elimination efforts, since it could facilitate identification of imported malaria cases.
I claim no ownership over the original source code, and all attribution and acknowledgement should be referred to the original project, and the associated publication1. I have simply provided this wrapper in the hope that it might be helpful for others, and have provided a tutorial for basic use.
-
Schaffner, S.F., Taylor, A.R., Wong, W. et al. hmmIBD: software to infer pairwise identity by descent between haploid genotypes. Malar J 17, 196 (2018). (https://malariajournal.biomedcentral.com/articles/10.1186/s12936-018-2349-7) ↩︎