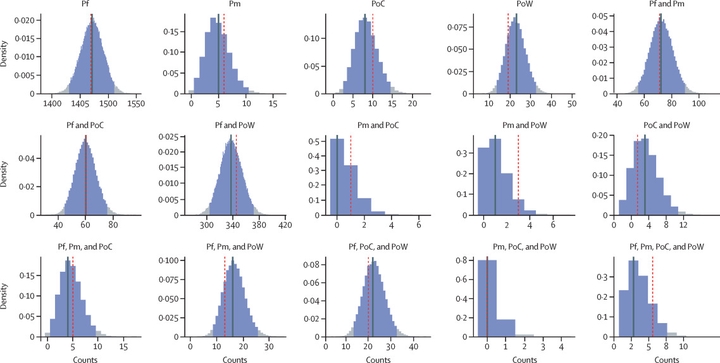
icer was designed to assess for interference between Plasmodium species to see if some species appeared in infections more often than expected under the assumption of independent random infection events, i.e. one species type is introduced from one infectious bite.
icer works out the most likely population frequency of each species and the most likely number of infection types in individual by comparison to the observed data. These are then used to test if our data can be explained by this distribution by comparing the observed data to the bootstrapped samples from the resultant multinomial distribution describing the probability of being infected with each infection compositon type. icer was used to assess for species interactions amongst non-falciparum malaria species in Kenya1.
-
H. M. Akala, O. J. Watson, K. K. Mitei, D. W. Juma, R. Verity, L. A. Ingasia, B. H. Opot, R. O. Okoth, G. C. Chemwor, J. A. Juma, E. W. Mwakio, N. Brazeau, A. C. Cheruiyot, R. A. Yeda, M. N. Maraka, C. O. Okello, D. P. Kateete, J. R. Managbanag, B. Andagalu, B. R. Ogutu, E. Kamau, Plasmodium interspecies interactions during a period of increasing prevalence of Plasmodium ovale in symptomatic individuals seeking treatment: an observational study. Lancet Microbe. 2, e141–e150 (2021). (https://www.thelancet.com/journals/lanmic/article/PIIS2666-5247(21)00009-4/fulltext) ↩︎